2.2 Evolution
BLAST works because evolution is happening. Biological sequences show complex patterns of similarity to one another. In this regard, they mirror the external morphologies of the organisms in which they reside. You'll notice that birds, for example, show natural groupings. You don't have to be a biologist to see that ducks, geese, and swans comprise a reasonably natural group called the waterfowl, and that the similarities between ducks and geese seem too great to explain by mere coincidence. Biological sequences are no different. After all, the reason why ducks look like ducks and geese look like geese is because of their genes. Many molecular biologists are convinced that understanding sequence evolution is tantamount to understanding evolution itself.
Sequences change over time due to three forces: mutation, natural selection, and genetic drift. If you use BLAST, it's important to understand these forces because they form the biological foundation of similarity searches. The biological and mathematical foundations aren't the same, and are sometimes at odds. You need to understand both theories in order to knowledgeably interpret the sequence alignments in a BLAST report.
2.2.1 Mutation
A mutation is simply a change in a DNA sequence. What causes mutation? Many chemicals and conditions damage DNA, so its sequence either changes or ceases to be recognizable. Mutagenic agents are often called carcinogens because cancer is caused by the accumulation of mutations in genes that control cell division. But even in a world without carcinogens there would still be mutation because the process of DNA replication isn't perfect. Every time a cell divides, it must duplicate its DNA. The human genome is about three billion letters long, and the error rate of DNA replication is about one error in every 300 million letters, so you can expect about 10 mutations per genome duplication. Genome size varies, as does the replication error rate, so don't take the 10 mutations per genome replication as any kind of biological truth. Human beings are composed of about a trillion cells, and you might take a moment now and consider just how much mutation is going on in your own body. Whatever that large number is, it's infinitesimal compared to what's happening in the biosphere as a whole.
What happens when a mutation occurs in the protein-coding portion of a gene? Because the DNA is mutated, the mRNA is also mutated. This in turn may lead to a different protein, but not necessarily, because the genetic code is degenerate. Take a look at an example for which you mutate just one letter in a coding sequence. If the mutation changed a codon from TTA to TTG, for example, the protein would be unchanged because both codons translate to the amino acid leucine. Such mutations are called silent, synonymous, or same-sense because they don't affect the protein sequence in any way. If the mutation changed a TTA to a TTT, however, the codon would code for a different amino acid, phenylalanine. Such substitutions are called mis-sense mutations. Molecular biologists will often classify mis-sense mutations into either conservative or nonconservative substitutions, depending on whether the two amino acids are chemically similar to one another. Leucine and phenylalanine are both hydrophobic amino acids, and such a substitution would be considered conservative. Bioinformaticists, however, give a more rigorous and quantifiable definition of conservative (see Chapter 4). If the TTA codon is mutated to TAA, the codon becomes a stop codon, which causes the ribosome to stop translating the mRNA. This represents the most destructive kind of mutation, and is called a non-sense mutation. Non-sense mutations cause translation to terminate prematurely, and the result is a truncated protein that may function partially, not function at all, or be poisonous to the cell.
Not all mutations substitute one nucleotide for another. Some mutations may insert or remove nucleotides. In addition, there are duplications, inversions, and other large-scale rearrangements that destroy genes or even fuse them together. Insertions and deletions are often destructive because they change the reading frame of translation if they aren't additions/subtractions of a multiple of three (a whole codon). After such a frame-shift mutation, there are usually several mis-sense mutations caused by the out-of-frame codons, and then a premature stop codon that was not previously in frame. Insertions and deletions are therefore usually as disruptive as mis-sense mutations.
What happens to an organism with mutations? It depends on a lot of factors. A mutation may have disastrous consequences, it might prove beneficial, or it might have no effect at all. To understand the forces that govern sequence evolution, let's take a close look at natural selection and genetic drift.
2.2.2 Natural Selection
The theory of natural selection was developed to explain why organisms look the way they do and why they seem to "fit" their environments so well. For example, why do giraffes have such long necks? Historically, there have been a lot of explanations, but we'll skip those debates and focus on the theory of natural selection because it is simple and fits the data well. The theory has only three assumptions.
There must be variation within a population.
The variation must be heritable.
There must be differential reproduction based on variation.
In the case of the giraffe ancestor, those individuals with slightly longer necks were at an advantage because they could reach leaves higher in the trees. This advantage translates to more surviving offspring, and since the variation is heritable, they too will tend to have longish necks. Now, within this population of longish necked pre-giraffes, there is still more variation, and the cycle of selecting for longer-necked individuals can persist until you have something that looks like a modern giraffe. People often look at the organisms today and think that their form is "complete." But all organisms are undergoing change from one generation to the next. When you look at a giraffe, try thinking about it as a particular form at a snapshot in time, on its way to something perhaps taller, or shorter, or with wings and horns and a penchant for breathing fire.
When Charles Darwin formulated the theory of natural selection, he had no idea about mutations, DNA, proteins, or the genetic code. The theory was based solely on observation; there was no known mechanism. In the last 50 years, the advances in molecular biology have revolutionized our understanding of natural selection. We now understand why there is variation and what is being selected for and against. The why is that variation exists at the DNA level (called alleles by geneticists). The what is differences in genes.
Consider how protein structure is selected for or against. What if a mutation causes an amino acid in the hydrophobic core of a protein to be changed to something hydrophilic? Well, it probably wouldn't fold the same way anymore because the hydrophobic core of the globular structure now has a part that wants to be in an aqueous environment. In most cases, changes in protein structure are unfavorable and therefore selected against; however, sometimes they result in altered function, which is favorable in certain conditions. Such is the case with sickle cell anemia. A charged amino acid (glutamate) is changed to a hydrophobic one (valine), causing altered protein interactions at the surface. Disease results when both alleles of the gene have this change, but it offers some protection against malaria when present in only one allele. As natural selection would predict, the sickle cell allele, and therefore sickle cell anemia, is prominent in certain parts of the world where malaria is common.
Several take-home messages are worth stating quite clearly. First, there is an inexhaustible source of variation because mutation is constantly happening. Natural selection isn't going to run out of variation. Evolution isn't going to stop. Second, a mutation can't be declared either good or bad on its own. Even a mutation that introduces a stop codon can be beneficial. Look at seedless oranges. It might seem an abomination of nature that they can't reproduce by themselves, but it is this very fact that makes humans breed them. To the seedless orange, genes that allow seeds to form are the kiss of death.
2.2.3 Genetic Drift
The interplay between mutation and natural selection that was just outlined makes a nice story. Like most stories, though, the truth is a lot more complicated. Reading the previous section, you may have concluded that natural selection is an all-powerful force, responsible for determining every nucleotide in a DNA sequence. In such a world, you would expect proteins to be perfectly functioning machines and the DNA sequences that encode them to be the best possible sequence for the job. This might be true in a mathematical model involving infinite population size and limitless generations, but the real biological world is a harsh place subject to happenstance. Even if the highly advantageous mutation enabling X-ray vision were to arise in some individual, it might not end up in the gene pool if that person thinks he's Superman and tries to stop a runaway train.
Darwin was not aware of how variation is transmitted from generation to generation; he didn't have the concept of genes. Genes were introduced by Gregor Mendel to explain how hereditary information is transmitted from one generation to the next. Combining Mendelian genetics and natural selection led to the field of population genetics, which is chiefly concerned with the changes in allele frequencies over time. Mathematical simulations show quite clearly that allele frequencies can change by purely random processes. This behavior is called genetic drift, and it's based on the fact that populations aren't infinitely large.
Let's demonstrate genetic drift with an example. For simplicity, let's ignore new mutations and just consider an anonymoussite that has no consequence in natural selection. Assume there are only 10 individuals in the population, and that 5 have a C at this position and 5 have a T. Keeping the population fixed, in the next generation, the allele frequencies may change to C=0.6 and T=0.4 due to a runaway train or, less spectacularly, sampling error. All things being equal, in the next generation, there's a greater chance that the C will increase and the T will decrease. If this trend continues for a few generations, the T's may disappear from the population entirely at which point the C allele is considered fixed in the population. Alleles can be fixed very rapidly if some individuals move away to form a new population. This is called the founder effect. As you can see, changes in allele frequencies don't require mutation or natural selection.
2.2.4 The Neutral Theory of Evolution
Molecular biology and the discovery of the genetic code had a profound effect on evolutionary biology. One shocking realization was that many sites for mutation?for example, the third position in a codon or a nucleotide in the middle of an intron (a term defined later), are expected to be invisible to natural selection. This led Motoo Kimura to propose the neutral theory of evolution. It was somewhat heretical when first proposed because it deemphasized the role of natural selection, but the theory states that the majority of sequence evolution is purely random, the product of mutation and drift.
Imagine what happens to a sequence as it accumulates random mutations over time. At first, the sequence is nearly identical to the original. If the rate of mutation is relatively consistent, you can count the number of mismatches to determine how much time has passed. This turns out to be very useful and forms the basis for determining the probability that a DNA sample matches a particular person, for example. Eventually, the number of mutations becomes so great that the sequence is no longer recognizably similar to the original. At this point, the sequence is saturated for mutation. Saturated sequences can't be used to measure time, but they are still very useful because they indicate which sequences aren't under selective pressure. By inference, those that remain similar over long periods of time are under selective pressure. As a practical example, when comparing the human and puffer fish genomes, you find that most of the conserved sequence is in genes.
One of the great debates of evolutionary biology is the relative importance of natural selection and neutral evolution in the formation of species. We don't need to be overly concerned with this argument because we're more interested in how sequences change over time, and for this we can observe actual sequence data.
2.2.5 Molecular Clocks
If you compare the sequences from related organisms, it is clear that certain positions don't change much over time while others change very rapidly. For example, parts of the ribosomal RNA are identical in every organism sequenced to date, from bacteria to humans. These subsequences are so important that if they change, the organism dies. Clearly, these are under intense selective pressure. There are other sites, such as third codon positions, that are only mildly affected by selection and tend to drift. There are even sequences, such as viral coat proteins, in which selection acts to promote variation, and these change very rapidly. Regardless of the underlying mechanism, it is possible to use the rate of change as a molecular clock.
If you know the mutation rate for a particular sequence, you can use it to determine how long ago two sequences diverged. Suppose you have the same protein sequence from both cats and dogs, and there are 10 differences between them. From the fossil record, you estimate that cats and dogs had a common ancestor 50 million years ago. Now when you compare the cat sequence to the same sequence in humans, you find 12 differences. You can now estimate that carnivores and humans shared a common ancestor 60 million years ago. We're using a very simple model here that treats all positions identically and we're not using real data, but this is the general idea behind molecular clocks.
The key to using molecular clocks is that the sequences must "tick" at the appropriate rate. The hypothetical protein in the last example is a poor choice for determining how long ago humans and chimps last shared a common ancestor because one difference here or there would lead to a large difference in the estimated time. Sequences that tick too fast are also not appropriate because they are prone to saturation.
2.2.6 Homology, Phylogeny, and Trees
When looking at the biological world around you, you see only what exists today. You can't get a clear picture of what the world looked like 100 million years ago. However, you can see relationships between organisms and make inferences. For example, you don't know what the last common ancestor of humans, chimpanzees, and gorillas looked like, but you can guess that it looked more like an ape than a bird. This is also the case at the sequence level; proteins from humans and chimps are much more similar to each other than either is to a bird. The study of relationships between organisms is called phylogenetics.
By definition, two sequences are homologous if they share a common ancestor. Two sequences are either homologous or they aren't. However, people often misuse the term and say something like "these two sequences are 80 percent homologous." What they usually mean is that two sequences are 80 percent identical and not that there is an 80 percent chance that they have a common ancestor. Determining if two sequences are indeed homologous requires making inferences. This isn't always a simple task; sometimes homology can be stated with near certainty, but not always. Sequences may appear to be related from chance similarity (or convergent evolution).
Sequence homology is further refined by the terms orthologous and paralogous. Sequences separated by speciation are called orthologs, while sequences separated by duplication are called paralogs. The genes for myoglobin in humans and mice are orthologs; they are the same gene in different species. If the myoglobin gene is duplicated in humans, the two myoglobins will be paralogs of each other. It's somewhat confusing, but both human paralogs would be considered orthologous to the mouse myoglobin. It is generally the case that the most similar genes between species are orthologs, and this is often used as an operational definition.
2.2.7 The Tree of Life
An introduction to molecular evolution would be incomplete without an overview of life on Earth. You may have learned in an introductory biology class that there are five taxonomic kingdoms (animals, plants, fungi, monera, and protista). This is based largely on what can be seen with your eyes or a microscope. Molecular biology opened up a new way to classify organisms based on sequences rather than external features. Figure 2-4 shows a tree for various organisms based on ribosomal DNA sequence. There are three obvious domains that Carl Woese called the Bacteria, Archaea, and Eucarya. Note that the arrow in the figure points to the root of the plants, animals, and fungi. From this perspective, the traditional five kingdoms are a bit nearsighted.
Figure 2-4. Tree of life based on rRNA sequence (Diagram courtesy of Norman Pace. Used with permission.)
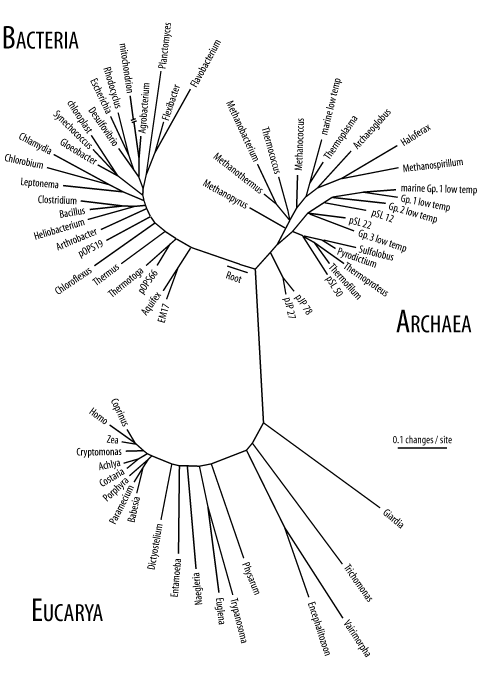
gIn terms of genomes and overall cell structure, there are only two major divisions: the prokaryotes (bacteria and archaea) and eukaryotes. Except in rare cases, prokaryotes are microscopic organisms that are usually shaped like rods or spheres. Some of the more famous prokaryotes include Escherichia coli (a bacterium that lives in your gut and is a favorite model organism for microbiologists) and Yersinia pestis (the bacterium that causes bubonic plague). The major distinguishing feature of prokaryotes is that DNA replication, transcription, and translation all take place in the same compartment of the cell because there is only one compartment in the cell.
Eukaryotes come in many shapes and sizes, primarily because they can form multi-cellular organisms such as birds and trees. But some eukaryotes are simple, single-celled organisms such as Saccharomyces cereviseae (the yeast used for making beer). All eukaryotes have a nucleus (karya is Greek for nucleus) in which DNA is stored, in addition to other membranous organelles. Interestingly, most eukaryotes contain mitochondria. These organelles have their own genome and are descended from bacteria that long ago entered a cooperative relationship with eukaryotes. This is also true of chloroplasts, which are responsible for photosynthesis in plants. It is thought that eukaryotes are a fusion of two bacteria, one a Eubacteria and one an Archaebacteria. So the next time you munch on a carrot, you might consider how many genomes are really in there.
So far, this chapter has neglected viruses. Where do they fit in? By most definitions, viruses aren't even alive; they don't grow or have repair processes. Viruses seem to break every rule of biology. Some viruses infect prokaryotes and others that parasitize eukaryotes. Viruses come in many different shapes and have wildly different lifestyles. Some have genomes made from RNA instead of DNA, and others have single-stranded rather than double-stranded genomes.







